August 1, 2025 - A long-standing collaboration with the research group of Bernd Rieger (TU Delft) has resulted in the publication of our new method to improve single localization microscopy.
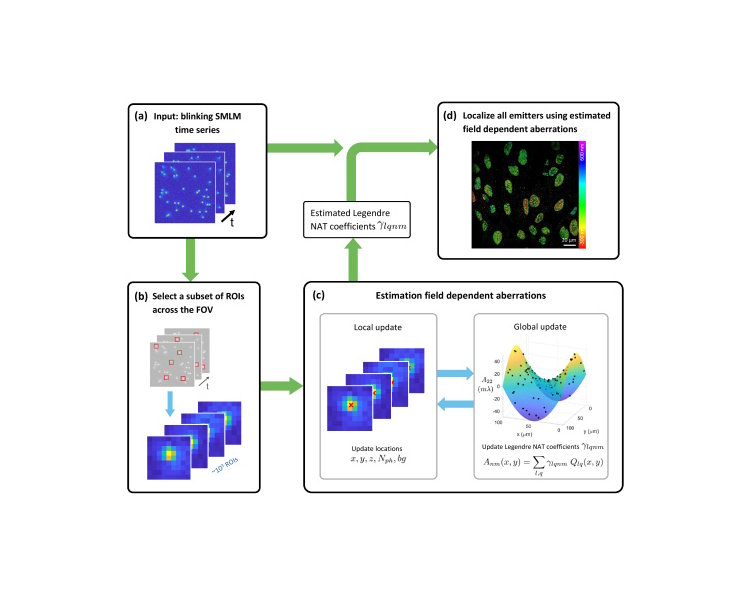
In super-resolution microscope imaging, scientists try to pinpoint the exact locations of single molecules. However, the images can become distorted depending on where in the view you’re looking, making it harder to get accurate results. Usually, fixing these distortions requires extra calibration steps with special samples. This study introduces a new method that can detect and correct these distortions automatically, using just the data from the regular imaging process without additional steps or materials needed. Stijn and I have been working on using GPUs to ensure the method works efficiently even on large images.
Abstract
Image quality in single-molecule localization microscopy depends largely on the accuracy and precision of the localizations. While under ideal imaging conditions, the theoretically obtainable precision and accuracy are achieved; in practice, this changes if (field-dependent) aberrations are present. Currently, there is no simple way to measure and incorporate these aberrations into the point-spread function (PSF) fitting; therefore, the aberrations are often taken as constant or neglected altogether. Here we introduce a model-based approach to estimate the field-dependent aberration directly from single-molecule data without a calibration step. This is made possible by using nodal aberration theory to incorporate the field dependency of aberrations into our fully vectorial PSF model. This results in a limited set of aberration fit parameters that can be extracted from the raw frames without a bead calibration measurement, also in retrospect. The software implementation is computationally efficient, enabling the fitting of a full 2D or 3D dataset within a few minutes. We demonstrate our method on 2D and 3D localization data of microtubuli, nuclear pore complexes, and nuclear lamina over fields of view of up to 180 µm and compare it with Gaussian fitting, spline-based fitting, and a deep-learning-based approach.
Citation
Isabel Droste, Erik Schuitema, Sajjad A. Khan, Myron Hensgens, Stijn Heldens, Carlas S. Smith, Ben van Werkhoven, Hylkje Geertsema, Keith A. Lidke, Sjoerd Stallinga, and Bernd Rieger “Calibration-free estimation of field-dependent aberrations for single-molecule localization microscopy across large fields of view” Optica 2025 https://doi.org/10.1364/OPTICA.560721